AWS HPC Blog
Enabling Rapid Genomic and Multiomic Data Analysis with Illumina DRAGEN™ v4.4 on Amazon EC2 F2 Instances
This post was contributed by Eric Allen (AWS), Olivia Choudhury (AWS), Mark Azadpour (AWS), Deepthi Shankar (Illumina), and Shyamal Mehtalia (Illumina)

The analysis of ever-increasing amounts of genomic and multiomic data demands efficient, scalable, and cost-effective computational solutions. Amazon Web Services (AWS) continues to support these workloads through FPGA accelerated compute offerings such as Amazon EC2 F2 instances.
Illumina’s DRAGEN (Dynamic Read Analysis for GENomics) secondary analysis solution has established itself as a leading secondary analysis solution for next generation sequencing data offering highly optimized algorithms for genomic analysis and hardware-accelerated implementations of comprehensive genomic and multiomic pipelines including germline DNA, somatic DNA, as well as bulk and single cell RNA analysis, proteomics, spatial, and more.
DRAGEN runs natively on EC2 F2 instances and provides customers with an accelerated approach for analyzing their biological data sets. Migration to F2s is streamlined as the same DRAGEN AMI is used for both F1 and F2, and analysis results will be equivalent1. In this post, we will discuss the performance characteristics of DRAGEN across different AWS compute environments as well as how to run the recently launched DRAGEN v4.4 on Amazon EC2 F2 instances.
Amazon EC2 F2 instances overview
F2 instances are the second generation of FPGA powered EC2 instances designed to accelerate analysis of genomic and multimedia data in the cloud. These instances offer significant improvements over their predecessors, the F1 instances, providing up to 60% better price performance. Here are the key features and specifications of F2 instances:
- FPGA Configuration: F2 instances are equipped with up to eight AMD Virtex UltraScale+ HBM VU47P FPGAs, each featuring 16GB of high-bandwidth memory (HBM).
- Processor: Powered by 3rd generation AMD EPYC (Milan) processors, F2 instances offer up to 192 vCPUs, which is three times the number of processor cores compared to F1 instances.
- Memory: These instances provide up to 2 TiB of system memory, doubling the memory capacity of F1 instances.
- Storage: F2 instances come with up to 7.6 TiB of NVMe SSD storage, which is twice the storage capacity of F1 instances.
- Networking: They offer up to 100 Gbps of networking bandwidth, quadrupling the network bandwidth available in F1 instances.
| Instance Name | FPGAs | vCPUs | Instance Memory | NVMe Storage | Network Bandwidth |
| f1.2xlarge | 1 | 8 | 122 | 470 | Up to 10 Gbps |
| f1.4xlarge | 2 | 16 | 244 | 940 | 10 Gbps |
| f2.6xlarge | 1 | 24 | 256 | 950 | 10 Gbps |
| F2.12xlarge | 2 | 48 | 512 | 1900 | 25 Gbps |
| f1.16xlarge | 8 | 64 | 976 | 4×940 | 25 Gbps |
| F2.48xlarge | 8 | 192 | 2,048 | 7,600 | 100 Gbps |
Table 1: Table showing comparative compute, memory, storage, and networking specs for F1 vs F2 instances.
Performance benchmarking approach
Illumina recommends using f1.4xlarge when using F1 instances and f2.6xlarge when using F2 instances. To assess performance across these instances, DRAGEN was configured and run on AWS following the Illumina DRAGEN user guide and the genome reference file links can be found on the Illumina DRAGEN Product Files web page
- Whole Genome Sequencing (WGS) analysis at approximately 35x depth using a publicly available sample. The sample HG002 from the NIST Genome in a Bottle project was used. This sample was analyzed using DRAGEN v4.4 Germline pipeline in two different manners. The first “basic” analysis included only basic alignment and small variant calling, to facilitate comparisons with common bioinformatics pipelines for germline samples, such as BWA/GATK. The second “full” analysis was with all variant callers (including for copy number and structural variants) and additional options such as pharmacogenetic star allele calling and HLA (Human Leukocyte Antigen) calling turned on, to produce a fully analyzed whole genome. In both cases, DRAGEN’s hg38 multigenome graph reference was used for WGS analysis. The sample data fastq files can be accessed on Amazon S3 using these links: fastq R1, fastq R2.
- Tumor Normal analysis using a pair of samples examined in a prior DRAGEN cancer analysis publication. These two samples had approximate depths of 110x (tumor) and 40x (normal). These samples were analyzed using the DRAGEN v4.4 Somatic pipeline, including alignment and small variant calling, plus CNV and SV analysis. DRAGEN’s hg38 linear genome reference was used for the Tumor Normal analysis. The sample data fastq files can be accessed on Amazon S3 using these links: Tumor fastq R1, Tumor fastq R2, Normal fastq R1, Normal fastq R2.
Speed & cost performance comparison: WGS analysis
WGS analysis using DRAGEN v4.4 demonstrated significant price performance advantages on Amazon EC2 F2 instances vs F1, while also generating equivalent analysis results between the two instance generations1:
- Basic WGS analysis, including alignment and small variant calling. DRAGEN v4.4 analysis on f2.6xlarge had 1.5x the speed and 40% of the EC2 compute cost vs f1.4xlarge.
- Complete WGS analysis, including alignment, small variant calling and calling of CNVs, SVs, and repeat expansions, and variant annotation. DRAGEN analysis on f2.6xlarge had 2x the speed and 30% of the EC2 compute cost vs f1.4xlarge.
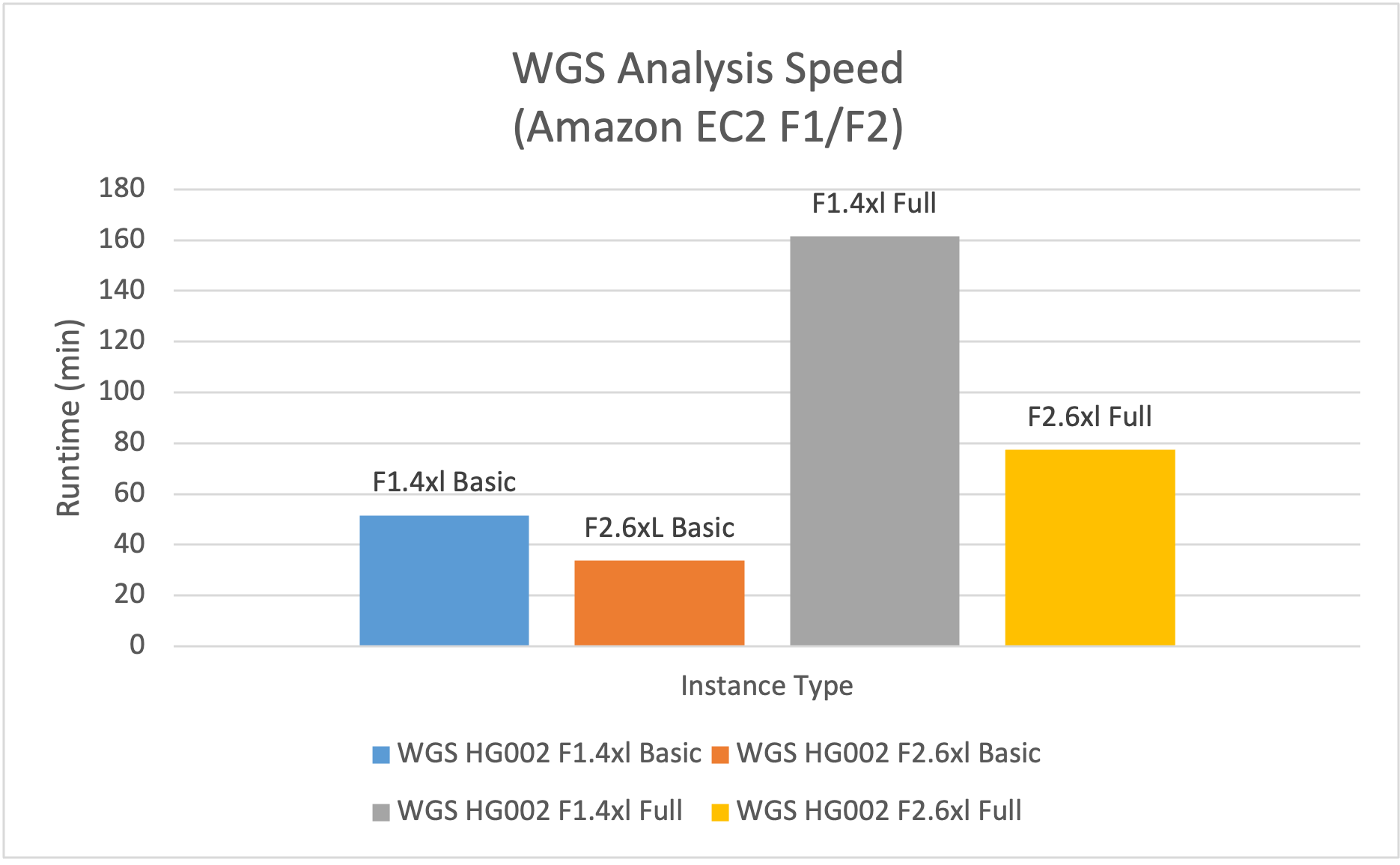
Figure 1: f2.6xlarge is 1.5x faster for WGS Basic Analysis and 2.1x faster for WGS Full Analysis than f1.4xlarge.
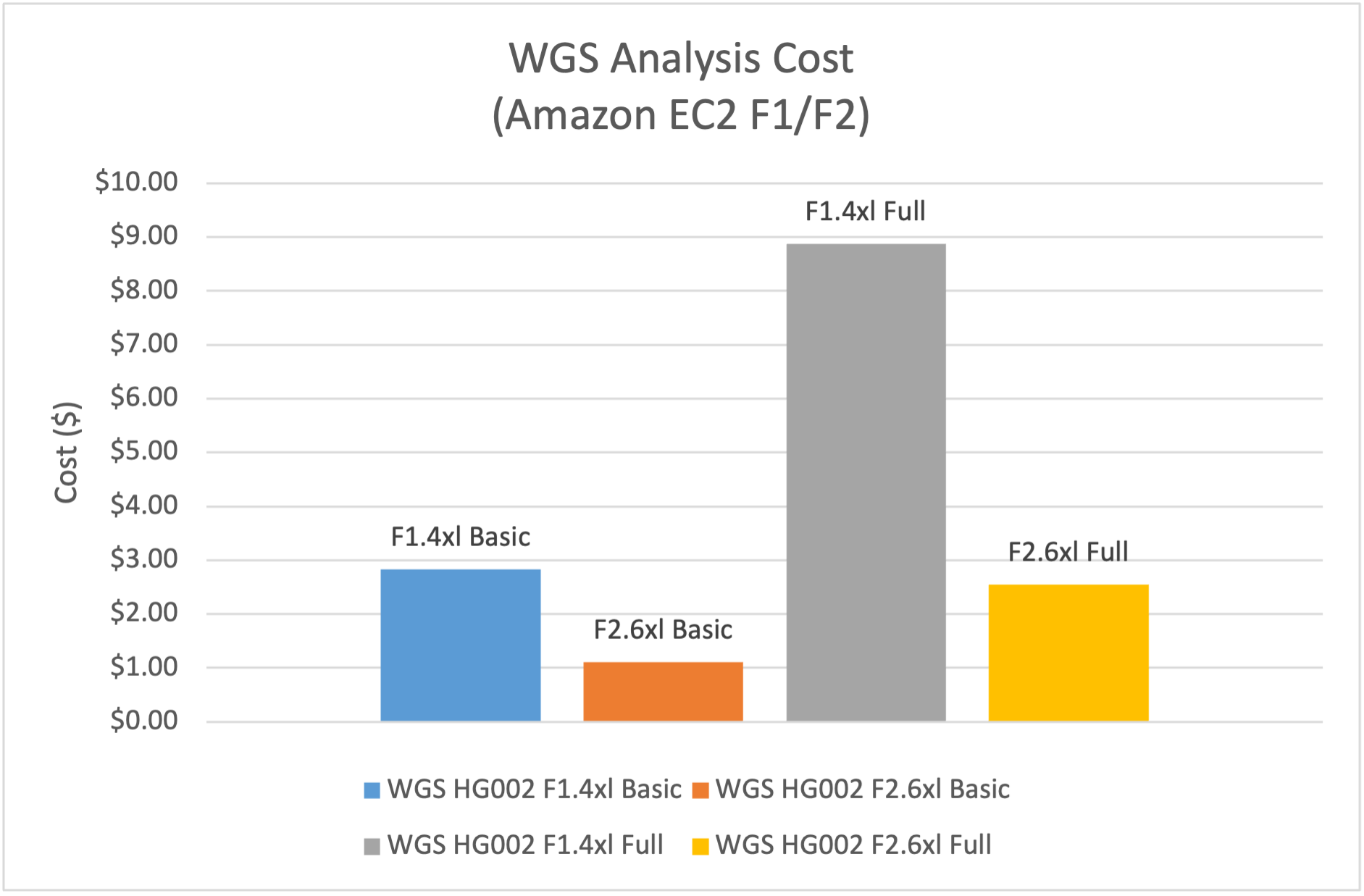
Figure 2: EC2 compute cost on f2.6xlarge is 40% of the cost on f1.4xlarge for WGS Basic Analysis
and 30% of the cost on f1.4xlarge for WGS Full Analysis.
Speed & cost performance comparison: Tumor Normal analysis
As with the WGS results, for Tumor Normal analysis DRAGEN v4.4 also demonstrated significant price performance advantages on Amazon EC2 F2 instances vs F1, while also generating equivalent analysis results between the two instance generations1:
- Tumor Normal analysis, including alignment, small variant calling, and calling of CNVs and SVs. DRAGEN analysis on f2.6xlarge had 1.7x the speed and 35% of the EC2 compute cost vs f1.4xlarge.
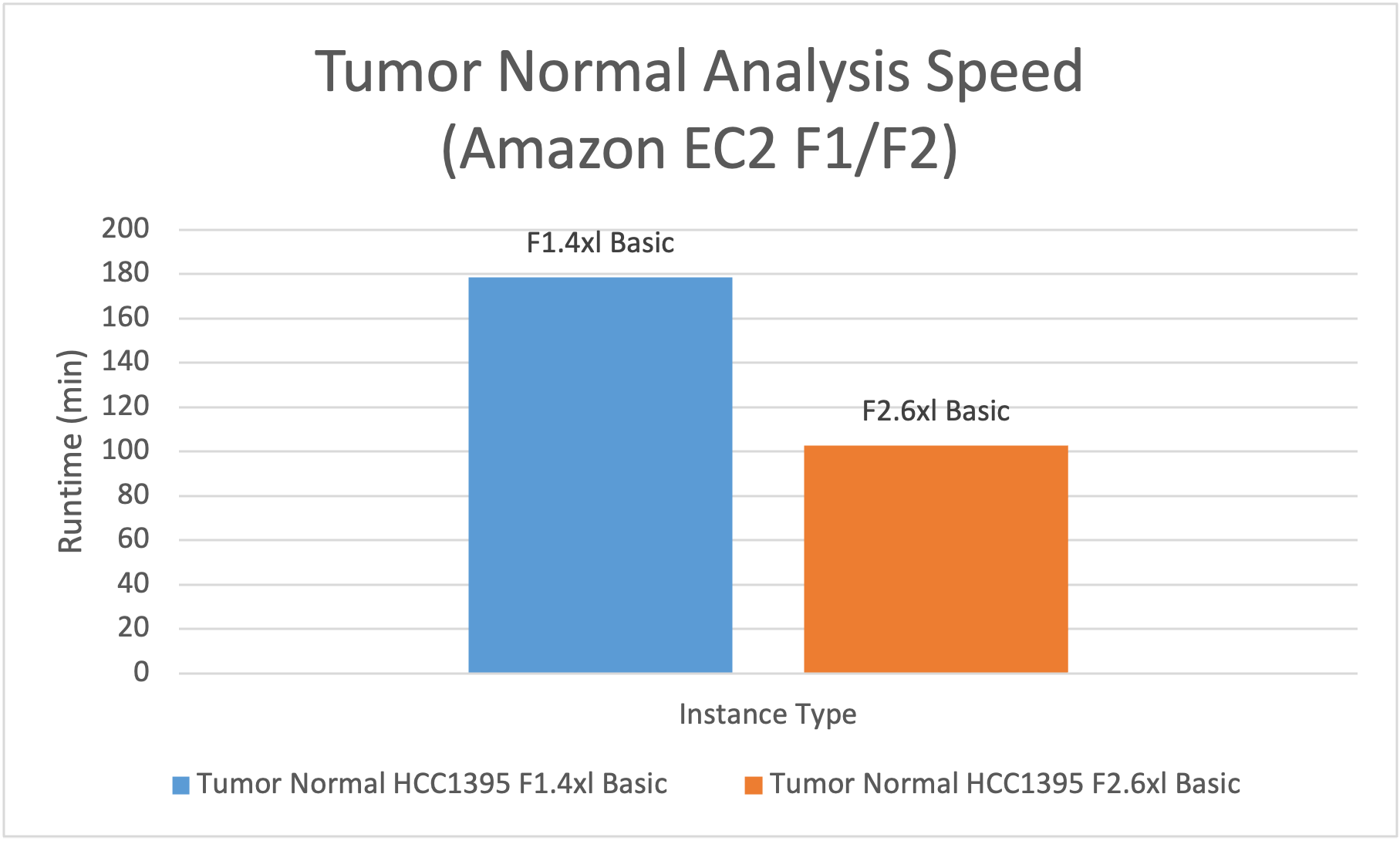
Figure 3: f2.6xlarge is 1.7x faster than f1.4xlarge for Tumor Normal Analysis.
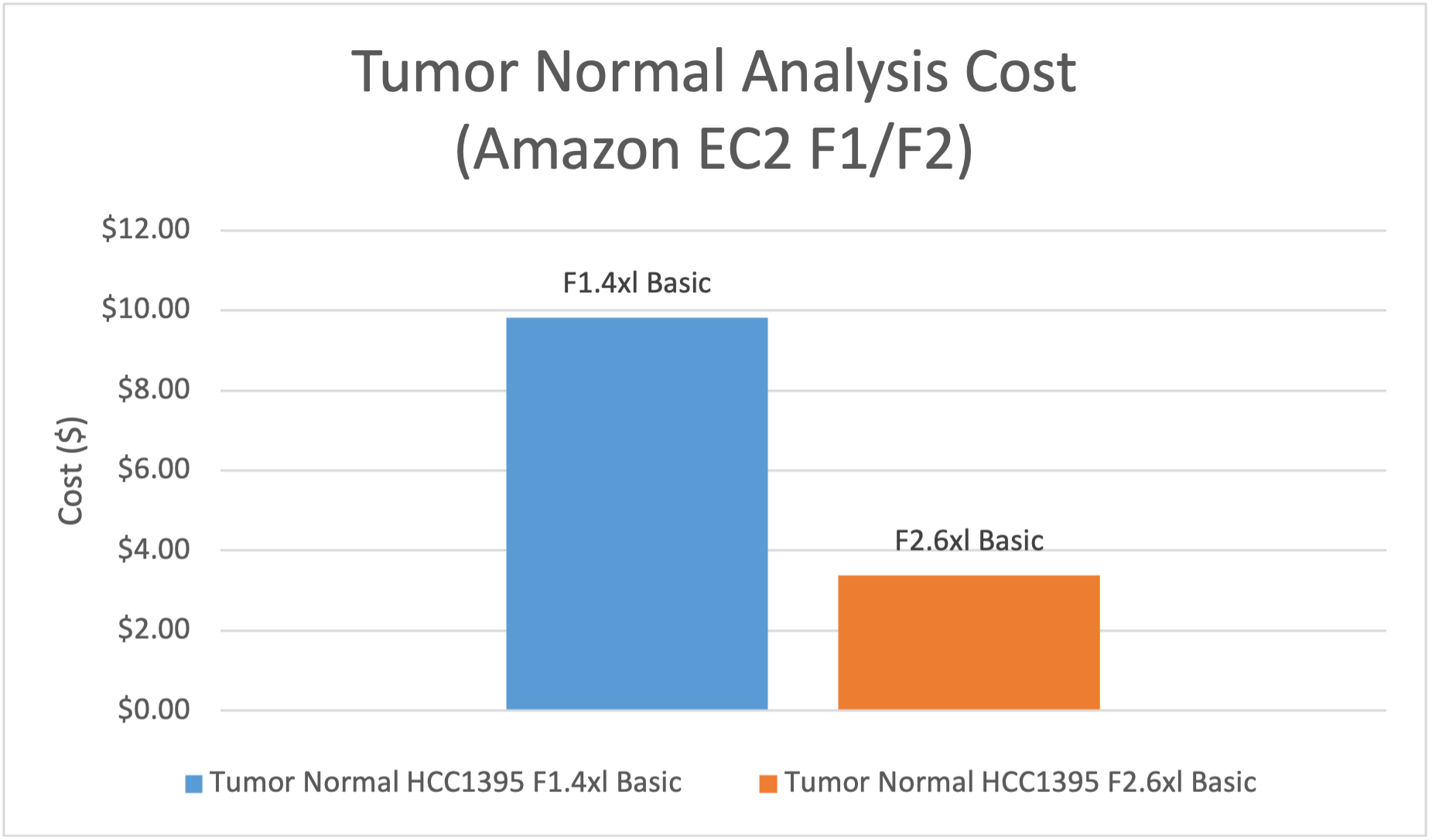
Figure 4: EC2 compute cost on f2.6xlarge is 35% of the cost on f1.4xlarge for WGS Tumor Normal Analysis.
Other advantages
Traditional genomic analysis pipelines using BWA-MEM and GATK running on CPUs have been the industry standard in the past, but DRAGEN has gained traction by providing speed and accuracy advantages. A number of peer reviewed publications have compared DRAGEN speed and accuracy vs BWA/GATK based pipelines running on CPU. For example, Ziegler et al (2022) found that DRAGEN analysis on FPGA hardware was over 8x faster and more accurate than BWA/GATK based pipelines running on CPU, while Sedlazek et al (2024) also found DRAGEN to have higher speed and accuracy vs BWA/GATK based pipelines.
Utilizing DRAGEN on F instances, which are powered by FPGAs, also offers power consumption advantages over traditional CPU and GPU solutions. FPGAs are inherently more energy-efficient, consuming less power for the same level of computational performance for these workloads. This is particularly important in genomic data analysis tasks, where the volume of data and amount of processing time required can be immense.
For instance, FPGAs achieve higher performance per watt compared to CPUs and GPUs for DRAGEN WGS workloads. FPGA-based accelerators can deliver superior throughput with lower power consumption. This is due to the dynamic customizability of FPGAs, which allows for optimized configurations that further enhance energy efficiency. In contrast, CPUs and GPUs, while powerful, often require more energy to perform the same tasks, leading to higher operational costs and a larger environmental footprint.
The lower power consumption of FPGAs translates to reduced cooling requirements, which can be a significant cost factor in large-scale computing environments. Additionally, the energy efficiency of FPGAs makes them an attractive option for high-performance computing applications, where scalability and cost-effectiveness are critical.
In summary, leveraging DRAGEN on Amazon EC2 ‘F’ family instances provides a more energy-efficient solution for genomic data analysis compared to traditional CPU or GPU-based approaches, offering both cost savings and environmental benefits.
F2 instance availability and cloud implementation options
Amazon EC2 F2 instances are available in multiple AWS Regions, including US East (N. Virginia), US West (Oregon), Europe (London), and Asia Pacific (Sydney), with plans for expanded Regional availability. They come in different sizes, such as f2.6xlarge, f2.12xlarge and f2.48xlarge, catering to various workload requirements.
When configuring storage and compute for running DRAGEN workloads on F instances on AWS, it’s important to select the right options to balance performance and cost. Consider storage options such as Amazon EBS gp3 volumes configured for RAID, Amazon FSx for Lustre for higher throughput, and Amazon Elastic File System (EFS) for persistent storage. Additionally, streaming BAM files and reference data from Amazon Simple Storage Service (Amazon S3) or using MountPoint for Amazon S3 to mount S3 buckets on the local file system can provide economical and simplified file access. By carefully choosing and configuring these storage solutions, you can ensure optimal performance and cost-effectiveness for your HPC workloads. Additionally, consider using Illumina Connected Analytics (ICA) or AWS Batch for workflow management. Illumina provides guidance for running DRAGEN on AWS as part of their online DRAGEN user guide.
Conclusion
In summary, Amazon EC2 F2 instances represent a significant advancement in FPGA-powered cloud computing, offering enhanced performance, memory, storage, and networking capabilities over the previous generations. The combination of DRAGEN’s comprehensive pipelines with Amazon EC2 F2’s improved computational power delivers faster, more efficient processing of complex datasets – from whole genome sequencing to single-cell RNA analysis.
Start your move to F2s today. Please reach out to your AWS account team or Illumina for help with your migration to AWS EC2 F2 instances.
For more info about DRAGEN on AWS, search for DRAGEN Complete Suite on AWS Marketplace, view the DRAGEN v4.4 blog, or visit Illumina’s Informatics Solutions home page.
References
Scheffler, K et al. “Somatic small-variant calling methods in Illumina DRAGEN™ Secondary Analysis.” BioRxiv (2023). https://www.biorxiv.org/content/10.1101/2023.03.23.534011v2
Sedlazech, FJ et al. “Comprehensive genome analysis and variant detection at scale using DRAGEN.” Nature (2024). https://www.nature.com/articles/s41587-024-02382-1
Ziegler, A et al. “Comparison of calling pipelines for whole genome sequencing: an empirical study demonstrating the importance of mapping and alignment.” Nature (2022). https://pubmed.ncbi.nlm.nih.gov/36513709/
Footnotes
- Equivalence between F1 and F2 hard-filtered.vcf and other results files based on analysis results using DRAGEN 4.4.4 following Illumina DRAGEN usage instructions. The vim diff command shows the only difference between the two VCF files is an entry showing the analysis execution time. Customers can do similar analyses themselves to verify or reach out to Illumina for more information.