AWS Big Data Blog
Medidata’s journey to a modern lakehouse architecture on AWS
This post was co-authored by Mike Araujo Principal Engineer at Medidata Solutions.
The life sciences industry is transitioning from fragmented, standalone tools towards integrated, platform-based solutions. Medidata, a Dassault Systèmes company, is building a next-generation data platform that addresses the complex challenges of modern clinical research. In this post, we show you how Medidata created a unified, scalable, real-time data platform that serves thousands of clinical trials worldwide with AWS services, Apache Iceberg, and a modern lakehouse architecture.
Challenges with legacy architecture
As the Medidata portfolio of data offerings expanded and our strategy evolved to offering an end-to-end clinical data platform experience, the team recognized the need to evolve the data platform architecture. The following diagram shows Medidata’s legacy extract, transform, and load (ETL) architecture.
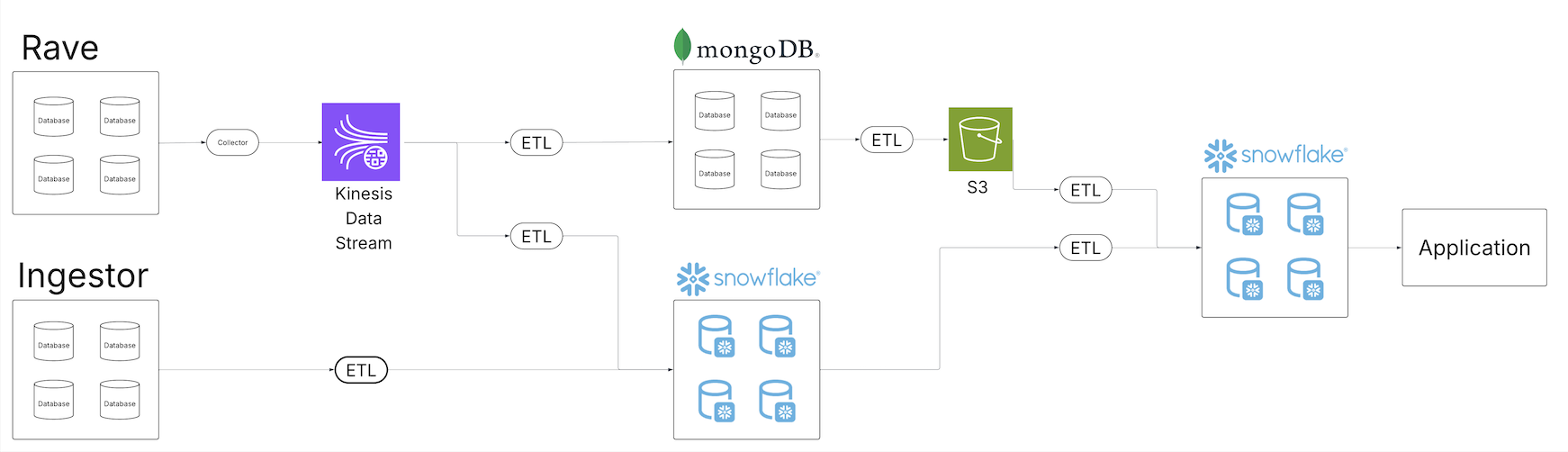
The legacy system was built on scheduled batch jobs, which served Medidata well for a long time, but wasn’t designed for a unified view of data across a growing ecosystem. Batch jobs ran at different intervals, requiring scheduling buffers to make sure upstream jobs completed on time. As data volume grew, these schedules became more complex, introducing latency between ingestion and processing. Different consumers pulling from various data services also meant teams were continuously building new pipelines for each delivery stack.
As the number of pipelines grew, so did the maintenance burden. More operations meant more potential points of failure and more complex recovery. The observability systems were handling a growing volume of operational data, making root cause analysis more involved. Scaling for increased data volume required coordinated changes across all our systems.
Additionally, having data pipelines and copies spread across different technologies and storage systems meant access controls had to be managed in multiple places. Propagating access control changes correctly across all systems added complexity for both consumers and producers.
Solution overview
With the advent of Clinical Data Studio (Medidata’s unified data management and analytics solution for clinical trials) and Data Connect (Medidata’s data solution for acquiring, transforming, and exchanging electronic health record (EHR) data across healthcare organizations), Medidata introduced a new world of data discovery, analysis, and integration to the life sciences industry powered by open source technologies and hosted on AWS. The following diagram illustrates the solution architecture.
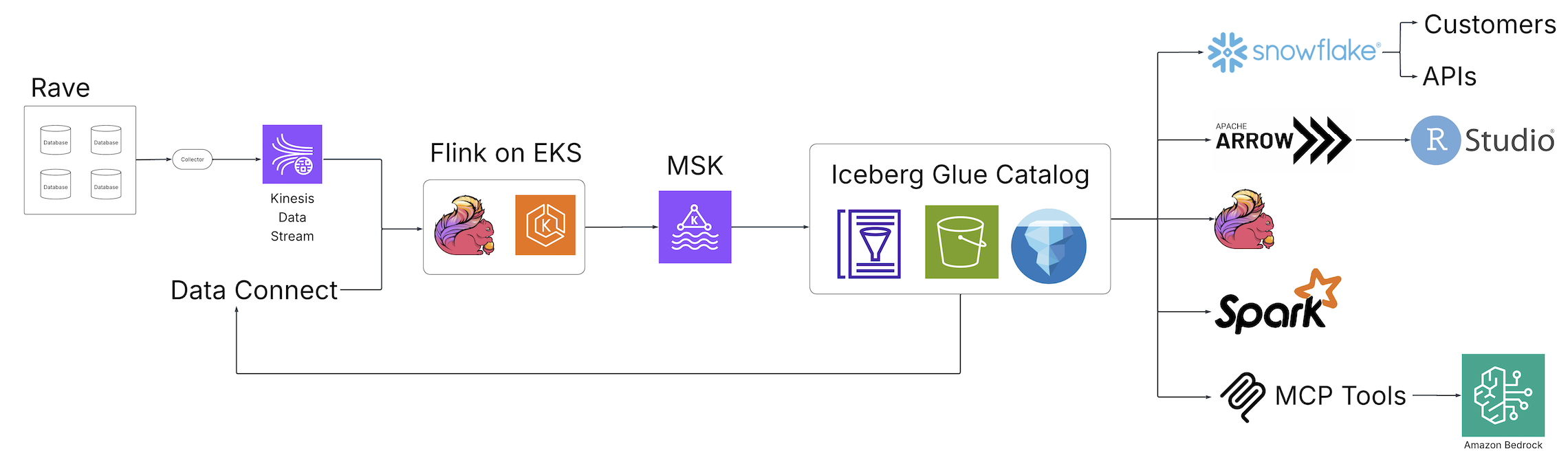
Fragmented batch ETL jobs were replaced by real-time Apache Flink streaming pipelines, an open source, distributed engine for stateful processing, and powered by Amazon Elastic Kubernetes Service (Amazon EKS), a fully managed Kubernetes service. The Flink jobs write to Apache Kafka running in Amazon Managed Apache Kafka (Amazon MSK), a streaming data service that manages Kafka infrastructure and operations, before landing in Iceberg tables backed by the AWS Glue Data Catalog, a centralized metadata repository for data assets. From this collection of Iceberg tables, a central, single source of data is now accessible from a variety of consumers without additional downstream processing, alleviating the need for custom pipelines to satisfy the requirements of downstream consumers. Through these fundamental architectural changes, the team at Medidata solved the issues presented by the legacy solution.
Data availability and consistency
With the introduction of the Flink jobs and Iceberg tables, the team was able to deliver a consistent view of their data across the Medidata data experience. Pipeline latency was reduced from numerous hours and sometimes days to minutes, helping Medidata customers realize a 99% performance gain from the data ingestion to the data analytics layers. Due to Iceberg’s interoperability, Medidata users saw the same view of the data regardless of where they viewed that data, minimizing the need for consumer-driven custom pipelines because Iceberg could plug into existing consumers.
Maintenance and durability
Iceberg’s interoperability provided a single copy of the data to satisfy their use cases, so the Medidata team could focus its observation and maintenance efforts on a five-times smaller subset of operations than previously required. Observability was enhanced by tapping into the various metadata components and metrics exposed by Iceberg and the Data Catalog. Quality management transformed from cross-system traces and queries to a single analysis of unified pipelines, with an added benefit of point in time data queries thanks to the Iceberg snapshot feature. Data volume increases are handled with out-of-box scaling supported by the entire infrastructure stack and AWS Glue Iceberg optimization features that include compaction, snapshot retention, and orphan file deletion, which provide a set-and-forget experience for solving a number of common Iceberg frustrations, such as the small file problem, orphan file retention, and query performance.
Security
With Iceberg at the center of its solution architecture, the Medidata team no longer had to spend the time building custom access control layers with enhanced security features at each data integration point. Iceberg on AWS centralizes the authorization layer using familiar systems such as AWS Identity and Access Management (IAM), providing a single and durable control for data access. The data also stays entirely within the Medidata virtual private cloud (VPC), further reducing the opportunity for unintended disclosures.
Conclusion
In this post, we demonstrated how legacy universe of consumer-driven custom ETL pipelines can be replaced with a scalable, high-performant streaming lakehouses. By putting Iceberg on AWS at the center of data operations, you can have a single source of data for your consumers.
To learn more about Iceberg on AWS, refer to Optimizing Iceberg tables and Using Apache Iceberg on AWS.